- R is a dialect of the S language
- S was initiated at the Bell Labs as an internal statistical analysis environment.
- Most well known implementation is S‐plus (most recent stable release was in 2010)
- R was first announced in 1993.
- The R core group was formed in 1997, who controls the source code of R (written in C)
- R 1.0.0 was released in 2000
- The current version is 3.5.0 (released on April 23, 2018)
- Previous version 3.4.4 was released on March 15, 2018
Programming in R
A brief introduction
Alex Pacheco
Research Computing
History
Features
- R is a dialect of the S language
- Language designed for statistical analysis
- Similar syntax
- Available on most platform/OS
- Rich data analysis functionalities and sophisticated graphical capabilities
- Active development and very active community
- CRAN: The Comprehensive R Archive Network
- Source code and binaries, user contributed packages and documentation
- More than 6,000 packages available on CRAN as of last week
- Free to use
Alternatives to R
- S-PLUS: commercial verison of S
- Gretl: open-source statistical package, mainly for econometrics
- SPSS: widely used program for statistical analysis in social science
- PSPP: free alternative to SPSS
- SAS: proprietary software that can be used with very large datasets such as census data
- STATA: proprietary software that is often used in economics and epidemiology
- MATLAB: proprietary software used widely in the mathematical sciences and engineering
- GNU Octave: free alternative to MATLAB
- Python: general programming language
Installing R
- R can be installed on Windows, Mac OSX and Linux from CRAN.
- R version 3.4.3 available on LUApps at https://luapps.lehigh.edu
Running R
- From Command Line on *NIX
- Enter
Ron the command line (if you have modified yourPATHcorrectly)
- Enter
- Batch Mode on *NIX
- Use the
Rscript filename.Rcommand to execute commands from a file,filename.R
- Use the
cat hello.R
## print("Hello World!")
Rscript hello.R
## [1] "Hello World!"
RStudio
- RStudio is the most popular (de facto) environment for running R on all platforms.
- free and open source IDE for R. Can be installed on Windows, Mac OSX and Linux.
- user interface comparable to other IDEs or software such as MATLAB.
- more suited for development
- Version 1.1.383 available on LUApps at https://luapps.lehigh.edu
Anaconda Python Distribution
- Anaconda Python distribution is the most popular platform for Python
- It provides
- a convenient install procedure for over 1400 Data Science libraries for Python and R
- conda to manage your packages, dependencies, and environments
- anaconda navigator: a desktop portal to install and launch applications and editors including Jupyter, RStudio, Visual Studio Code, and Spyder
- install r-essentials from anaconda navigator or
conda install r-essentialsfrom the command line
- install r-essentials from anaconda navigator or
- Jupyter Notebooks is an alternative to RStudio for writing scripts and
workflows that you can share with others.
- It is ideal for reproducible research or data reporting
- Visit https://go.lehigh.edu/linux to use R, Anaconda and other Linux software installed and maintained by the Research Computing group on your local Linux laptop or workstation
- Click here for instructions to run Jupyter Notebooks on Sol
Get Started with R
- Use the console to use R as a simple calculator
1 + 2
## [1] 3
- The assignment symbol is "<-". The classical "=" symbol can also be used
a=2+3
b<-10/a
a
## [1] 5
b
## [1] 2
Get Started with R
- install packages from CRAN, for e.g. knitr
install.packages('knitr')
- load a library, for e.g. knitr
library(knitr)
- Help from command line
?<command name>
??<part of command name/topic>
or search in the help page in RStudio
getwd(): display current working directorysetwd('dir'): change current working director todir
Data Classes
- R has five atomic classes
- Numeric
- Double is equivalent to numeric.
- Numbers in R are treated as numeric unless specified otherwise.
- Integer
- Complex
- Character
- Logical
- TRUE or FALSE
- You can convert data from one type to the other using the
as.<Type>functions - To check the class of an object, use the
is.<Type>function.
Example
a <- 3
b <- sqrt(a)
b
## [1] 1.732051
c <- 2i
d <- TRUE
d
## [1] TRUE
as.numeric(d); as.character(b); is.complex(c)
## [1] 1
## [1] "1.73205080756888"
## [1] TRUE
Data Objects‐ Vectors
- Vectors can only contain elements of the same class
- Vectors can be constructed by
- Using the
c()function (concatenate)
- Using the
- Coercion will occur when mixed objects are passed to the
c()function, as if theas.<Type>()function is explicitly called- Using the
vector()function
- Using the
- One can use
[index]to access individual element- Indices start from 1
Examples
# "#" indicates comment
# "<-" performs assignment operation (you can use "=" as well, but "<-" is preferred)
# numeric (double is the same as numeric)
d <- c(1,2,3)
# character
d <- c("1","2","3")
# you can covert at object with as.TYPE
# as. numeric changes the character vector created above to numeric
as.numeric(d)
## [1] 1 2 3
# The conversion doesn't always work though
as.numeric("a")
## Warning: NAs introduced by coercion
## [1] NA
Examples (contd)
x <- c(0.5, 0.6) ## numeric
x <- c(TRUE, FALSE) ## logical
x <- c(T, F) ## logical
x <- c("a", "b", "c") ## character
# The ":" operator can be used to generate integer sequences
x <- 9:29 ## integer
x <- c(1+0i, 2+4i) ## complex
x <- vector("numeric", length = 10)
# Coercion will occur when objects of different classes are mixed
y <- c(1.7, "a") ## character
y <- c(TRUE, 2) ## numeric
y <- c("a", TRUE) ## character
# Can also coerce explicitly
x <- 0:6
class(x)
## [1] "integer"
as.logical(x)
## [1] FALSE TRUE TRUE TRUE TRUE TRUE TRUE
Vectorized Operations
- Lots of R operations process objects in a vectorized way
- more efficient, concise, and easier to read.
x <- 1:4; y <- 6:9
x + y
## [1] 7 9 11 13
x > 2
## [1] FALSE FALSE TRUE TRUE
x * y
## [1] 6 14 24 36
print( x[x >= 3] )
## [1] 3 4
Data Objects - Matrices
- Matrices are vectors with a dimension attribute
- R matrices can be constructed
- Using the
matrix()function- Passing an dim attribute to a vector
- Using the
cbind()orrbind()functions
- Using the
- R matrices are constructed column‐wise
- One can use
[<index>,<index>]to access individual element
Example
# Create a matrix using the matrix() function
m <- matrix(1:6, nrow = 2, ncol = 3)
m
## [,1] [,2] [,3]
## [1,] 1 3 5
## [2,] 2 4 6
dim(m)
## [1] 2 3
attributes(m)
## $dim
## [1] 2 3
Example
# Pass a dim attribute to a vector
m <- 1:10
m
## [1] 1 2 3 4 5 6 7 8 9 10
dim(m) <- c(2, 5)
m
## [,1] [,2] [,3] [,4] [,5]
## [1,] 1 3 5 7 9
## [2,] 2 4 6 8 10
Example
# Row binding and column binding
x <- 1:3
y <- 10:12
cbind(x, y)
## x y
## [1,] 1 10
## [2,] 2 11
## [3,] 3 12
rbind(x, y)
## [,1] [,2] [,3]
## x 1 2 3
## y 10 11 12
Example
# Slicing
m
## [,1] [,2] [,3] [,4] [,5]
## [1,] 1 3 5 7 9
## [2,] 2 4 6 8 10
# element at 2nd row, 3rd column
m[2,3]
## [1] 6
# entire i<sup>th</sup> row of m
m[2,]
## [1] 2 4 6 8 10
# entire j<sup>th</sup> column of m
m[,3]
## [1] 5 6
Data Objects - Lists
- Lists are a special kind of vector that contains objects of different classes
- Lists can be constructed by using the
list()function - Lists can be indexed using
[[ ]]
# Use the list() function to construct a list
x <- list(1, "a", TRUE, 1 + 4i)
x
## [[1]]
## [1] 1
##
## [[2]]
## [1] "a"
##
## [[3]]
## [1] TRUE
##
## [[4]]
## [1] 1+4i
Data Objects - Data Frames
- Data frames are used to store tabular data
- They are a special type of list where every element of the list has to have the same length
- Each element of the list can be thought of as a column
- Data frames can store different classes of objects in each column
- Data frames also have a special attribute called
row.names - Data frames are usually created by calling
read.table()orread.csv() - Can be converted to a matrix by calling
data.matrix()
Names
- R objects can have names
# Each element in a vector can have a name
x <- 1:3
names(x)
## NULL
names(x) <- c("a","b","c")
names(x)
## [1] "a" "b" "c"
x
## a b c
## 1 2 3
Names (contd)
# Lists
x <- list(a = 1, b = 2, c = 3)
x
## $a
## [1] 1
##
## $b
## [1] 2
##
## $c
## [1] 3
# Names can be used to refer to individual element
x$a
## [1] 1
Names (contd)
# Columns and rows of matrices
m <- matrix(1:4, nrow = 2, ncol = 2)
dimnames(m) <- list(c("a", "b"), c("c", "d"))
m
## c d
## a 1 3
## b 2 4
Querying Object Attributes
- The
class()function - The
str()function - The
attributes()function reveals attributes of an object (does not work with vectors)- Class
- Names
- Dimensions
- Length
- User defined attributes
- They work on all objects (including functions)
Example
m <- matrix(1:10, nrow = 2, ncol = 5)
str(matrix)
## function (data = NA, nrow = 1, ncol = 1, byrow = FALSE, dimnames = NULL)
str(m)
## int [1:2, 1:5] 1 2 3 4 5 6 7 8 9 10
str(str)
## function (object, ...)
Data Class - Factors
- Factors are used to represent categorical data.
- Factors can be unordered or ordered.
- Factors are treated specially by modelling functions like
lm()andglm()
# Use the factor() function to construct a vector of factors
# The order of levels can be set by the levels keyword
x <- factor(c("yes", "yes", "no", "yes", "no"), levels = c("yes", "no"))
x
## [1] yes yes no yes no
## Levels: yes no
Date and Time
- R has a Date class for date data while times are represented by POSIX formats
- One can convert a text string to date using the
as.Date()function - The
strptime()function can deal with dates and times in different formats. - The package "
lubridate" provides many additional and convenient features
# Dates are stored internally as the number of days since 1970-01-01
x <- as.Date("1970-01-01")
x
## [1] "1970-01-01"
as.numeric(x)
## [1] 0
x+1
## [1] "1970-01-02"
Data and Time (contd)
# Times are stored internally as the number of seconds since 1970-01-01
x <- Sys.time() ; x
## [1] "2018-03-22 08:40:06 EDT"
as.numeric(x)
## [1] 1521722406
p <- as.POSIXlt(x)
names(unclass(p))
## [1] "sec" "min" "hour" "mday" "mon" "year" "wday"
## [8] "yday" "isdst" "zone" "gmtoff"
p$sec
## [1] 6.479255
Missing Values
- Missing values are denoted by
NAorNaNfor undefined mathematical operations.is.na()is used to test objects if they areNAis.nan()is used to test forNaNNAvalues have a class also, so there are integerNA, characterNA, etc.- A
NaNvalue is alsoNAbut the converse is not true
x <- c(1,2, NA, 10,3)
is.na(x)
## [1] FALSE FALSE TRUE FALSE FALSE
is.nan(x)
## [1] FALSE FALSE FALSE FALSE FALSE
Missing Values (contd)
x <- c(1,2, NaN, NA,4)
is.na(x)
## [1] FALSE FALSE TRUE TRUE FALSE
is.nan(x)
## [1] FALSE FALSE TRUE FALSE FALSE
Arithmetic Functions
| Function | Description |
|---|---|
exp() | Exponentiation |
log() | Natural Logarithm |
log10() | Logarithm to base 10 |
sqrt() | square root |
abs() | absolute value |
sin() | sine |
cos() | cosine |
floor() | |
ceiling() | rounding of numbers |
round() |
Simple Statistic Functions
| Function | Description |
|---|---|
min() | minimum value |
max() | maximum value |
which.min() | location of minimum |
which.max() | location of maximum |
pmin() | elementwise minima of several vectors |
pmax() | elementwise maxima of several vectors |
sum() | sum of elements of a vector |
mean() | mean of elements of a vector |
prod() | products of elements of a vector |
Distributions and Random Variables
- For each distribution R provides four functions: density (
d), cumulative density (p), quantile (q), and random generation (r)- The function name is of the form
[d|p|q|r]<name of distribution> - e.g.
qbinom()gives the quantile of a binomial distribution
- The function name is of the form
| Distribution | Distribution name in R |
|---|---|
| Uniform | unif |
| Binomial | binom |
| Poisson | pois |
| Geometric | geom |
| Gamma | gamma |
| Normal | norm |
| Log Normal | lnorm |
| Exponential | exp |
| Student’s t | t |
Examples: Distributions and Random Variables
# Random generation from a uniform distribution.
runif(10, 2, 4)
## [1] 2.080494 2.140020 3.279643 2.292516 2.121182 2.465787 2.891568
## [8] 3.221651 2.835845 3.554286
# You can name the arguments in the function call.
runif(10, min = 2, max = 4)
## [1] 3.864708 3.558662 3.011422 2.551474 2.215152 3.845879 2.237049
## [8] 2.028424 2.695853 3.390850
# Given p value and degree of freedom, find the t-value.
qt(p=0.975, df = 8)
## [1] 2.306004
# The inverse of the above function call
pt(2.306, df = 8)
## [1] 0.9749998
User Defined Functions
- Similar to other languages, functions in R are defined by using the
function()directives - The return value is the last expression in the function body to be evaluated.
- Functions can be nested
- Functions are R objects
- For example, they can be passed as an argument to other functions
newDef <- function(a,b)
{
x = runif(10,a,b)
mean(x)
}
newDef(-1,1)
## [1] 0.1779019
Control Structures
- Control structures allow one to control the flow of execution.
if … else | testing a condition |
for | executing a loop (with fixed number of iterations) |
while | executing a loop when a condition is true |
repeat | executing an infinite loop |
break | breaking the execution of a loop |
next | skipping to next iteration |
return | exit a function |
Testing conditions
# Comparisons: <,<=,>,>=,==,!=
# Logical operations: !, &&, ||
if(x > 3 && x < 5) {
print ("x is between 3 and 5")
} else if(x <= 3) {
print ("x is less or equal to 3")
} else {
print ("x is greater or equal to 5")
}
For Loops
x <- c("a", "b", "c", "d")
# These loops have the same effect
# Loop through the indices
for(i in 1:4) {
print(x[i])
}
## [1] "a"
## [1] "b"
## [1] "c"
## [1] "d"
# Loop using the seq_along() function
for(i in seq_along(x)) {
print(x[i])
}
## [1] "a"
## [1] "b"
## [1] "c"
## [1] "d"
For Loops (contd)
# Loop through the name
for(letter in x) {
print(letter)
}
## [1] "a"
## [1] "b"
## [1] "c"
## [1] "d"
for(i in 1:4) print(x[i])
## [1] "a"
## [1] "b"
## [1] "c"
## [1] "d"
while loops
- The
whileloop can be used to repeat a set of instructions - It is often used when you do not know in advance how often the instructions will be executed.
- The basic format for a
whileloop iswhile(cond) expr
sum <- 1
while ( sum < 11 )
{
sum <- sum + 2;
print(sum);
}
## [1] 3
## [1] 5
## [1] 7
## [1] 9
## [1] 11
sum <- 12
while (sum < 11 )
{
sum <- sum + 2;
print(sum);
}
repeat loops
- The
repeatloop is similar to thewhileloop. - The difference is that it will always begin the loop the first time. The
whileloop will only start the loop if the condition is true the first time it is evaluated. - Another difference is that you have to explicitly specify when to stop the loop using the
breakcommand.
sum <- 1
repeat
{
sum <- sum + 2;
print(sum);
if (sum > 11)
break;
}
## [1] 3
## [1] 5
## [1] 7
## [1] 9
## [1] 11
## [1] 13
sum <- 12
repeat
{
sum <- sum + 2;
print(sum);
if (sum > 11)
break;
}
## [1] 14
break and next statements
- The
breakstatement is used to stop the execution of the current loop.- It will break out of the current loop.
- The
nextstatement is used to skip the statements that follow and restart the current loop.- If a
forloop is used then thenextstatement will update the loop variable.
- If a
x <- rnorm(5)
for(lupe in x)
{
if (lupe > 2.0)
next
if( (lupe<0.6) && (lupe > 0.5))
break
cat("The value of lupe is ",lupe,"\n");
}
## The value of lupe is 1.252302
## The value of lupe is -0.3448712
## The value of lupe is -0.1833472
## The value of lupe is 1.066976
## The value of lupe is -0.4986212
The apply Function
- The
apply()function evaluate a function over the margins of an array- More concise than the for loops (not necessarily faster)
# X: array objects
# MARGIN: a vector giving the subscripts which the function will be applied over
# FUN: a function to be applied
str(apply)
## function (X, MARGIN, FUN, ...)
x <- matrix(rnorm(200), 20, 10)
# Row means
apply(x, 1, mean)
## [1] -0.56706372 0.29605236 -0.12243105 0.39508406 0.48143361
## [6] 0.04930200 -0.20466170 0.12974536 -0.13387390 -0.02735853
## [11] 0.21488576 -0.33514770 0.11733424 0.65669985 0.53110367
## [16] -0.08488930 0.30794310 0.09569795 0.12904018 0.03674856
The apply Function (contd)
# Column sums
apply(x, 2, sum)
## [1] 0.1150799 3.3847897 4.1670340 8.4534105 -0.1794458 3.2395951
## [7] -0.3983110 -1.2475295 2.4118395 -0.2900143
# 25th and 75th Quantiles for rows
apply(x, 1, quantile, probs = c(0.25, 0.75))
## [,1] [,2] [,3] [,4] [,5] [,6]
## 25% -1.12981396 0.05320533 -0.76026179 -0.280979 0.1561278 -0.4971189
## 75% -0.03635382 0.65380948 -0.05765654 1.439553 0.7192037 0.5006148
## [,7] [,8] [,9] [,10] [,11] [,12]
## 25% -0.4871022 -0.4261296 -1.0766658 -0.6012603 -0.3460629 -0.8672751
## 75% 0.3523635 0.7871360 0.3282589 0.6461509 0.5666012 0.1798698
## [,13] [,14] [,15] [,16] [,17] [,18]
## 25% -0.695670 0.1628591 0.1135125 -0.5335172 -0.4598713 -0.5215956
## 75% 0.814849 1.3024599 1.1476166 0.3738642 1.1652113 0.6152672
## [,19] [,20]
## 25% -0.3562603 -0.8098604
## 75% 0.6595250 0.6965057
The apply Function (contd)
dim(x)
## [1] 20 10
# Change the dimensions of x
dim(x) <- c(2,2,50)
# Take average over the first two dimensions
apply(x, c(1, 2), mean)
## [,1] [,2]
## [1,] 0.04115467 0.1095874
## [2,] 0.21407873 0.0283082
rowMeans(x, dims = 2)
## [,1] [,2]
## [1,] 0.04115467 0.1095874
## [2,] 0.21407873 0.0283082
Other Apply Functions
lapply: Loop over a list and evaluate a function on each elementsapply: Same as lapply but try to simplify the resulttapply: Apply a function over subsets of a vectormapply: Multivariate version of lapply
R for Data Science
- The
tidyverseis a collection of R packages developed by RStudio’s chief scientist Hadley Wickham.ggplot2for data visualisation.dplyrfor data manipulation.tidyrfor data tidying.readrfor data import.purrrfor functional programming.tibblefor tibbles, a modern re-imagining of data frames.
- These packages work well together as part of larger data analysis pipeline.
- To learn more about these tools and how they work together, read R for Data Science.
Tidyverse
- What is Tidy Data?
- "Tidy data" is a term that describes a standardized approach to structuring datasets to make analyses and visualizations easier.
- The core tidy data principles
- Variable make up the columns
- Observations make up the rows
- Values go into cells
library(tidyverse)will load the core tidyverse packages:
library(tidyverse)
## ── Attaching packages ────────────────────────────────── tidyverse 1.2.1 ──
## ✔ tibble 1.4.2 ✔ purrr 0.2.4
## ✔ tidyr 0.8.0 ✔ dplyr 0.7.4
## ✔ readr 1.1.1 ✔ stringr 1.2.0
## ✔ tibble 1.4.2 ✔ forcats 0.2.0
## ── Conflicts ───────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::arrange() masks plyr::arrange()
## ✖ purrr::compact() masks plyr::compact()
## ✖ dplyr::count() masks plyr::count()
## ✖ dplyr::failwith() masks plyr::failwith()
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::id() masks plyr::id()
## ✖ dplyr::lag() masks stats::lag()
## ✖ dplyr::mutate() masks plyr::mutate()
## ✖ dplyr::rename() masks plyr::rename()
## ✖ dplyr::summarise() masks plyr::summarise()
## ✖ dplyr::summarize() masks plyr::summarize()
library(lubridate)
##
## Attaching package: 'lubridate'
## The following object is masked from 'package:plyr':
##
## here
## The following object is masked from 'package:base':
##
## date
Tidyverse
- Packages that are part of tidyverse but not loaded automatically
lubridatefor dates and date-timesmagrittrprovides the pipe, %>% used throughout the tidyverse.readxlfor .xls and .xlsx sheets.havenfor SPSS, Stata, and SAS data.
- packages that are not in the tidyverse, but are tidyverse-adjacent. They are very useful for importing data from other sources:
jsonlitefor JSON.xml2for XML.httrfor web APIs.rvestfor web scraping.DBIfor relational databases
Readr Package
readris to provide a fast and friendly way to read rectangular data (like csv, tsv, and fwf).readrsupports seven file formats with seven read_ functions:read_csv(): comma separated (CSV) filesread_csv2(): semicolon separated file and "," for decimal pointread_tsv(): tab separated filesread_delim(): general delimited filesread_fwf(): fixed width filesread_table(): tabular files where colums are separated by white-space.read_log(): web log files
- Usage
read_delim(file,delim)file: path to a file, a connection, or literal data
Example
# read daily usage report for Sol in AY 2016-17
# usage is reported in terms of SUs used and jobs submitted for
# serial (1 cpu), single or smp ( > 1 cpu but max of 1 node) and
# parallel or multi node (> 1 node) jobs
daily <- read_delim('http://webapps.lehigh.edu/hpc/training/soldaily1617-public.csv',delim=";")
## Parsed with column specification:
## cols(
## Type = col_character(),
## Name = col_character(),
## Department = col_character(),
## PI = col_character(),
## PIDept = col_character(),
## Status = col_character(),
## Day = col_date(format = ""),
## SerialJ = col_character(),
## Serial = col_character(),
## SingleJ = col_character(),
## Single = col_character(),
## MultiJ = col_character(),
## Multi = col_character(),
## TotalJ = col_character(),
## Total = col_character()
## )
#daily <- read_delim('http://webapps.lehigh.edu/hpc/training/soldaily1718-public.csv',delim=";")
- the
readrfunctions will just work: you supply the path to a file and you get atibbleback
Tibble
- A
tibble, ortbl_df, is a modern reimagining of the data.frame,- keeping what time has proven to be effective, and
- throwing out what is not.
Tibbles are data.frames that are lazy and surly
Create a tibble from an existing object with
as_tibble()create a new tibble from column vectors with
tibble()
tibble(x = 1:5, y = 1, z = x ^ 2 + y)
## # A tibble: 5 x 3
## x y z
## <int> <dbl> <dbl>
## 1 1 1.00 2.00
## 2 2 1.00 5.00
## 3 3 1.00 10.0
## 4 4 1.00 17.0
## 5 5 1.00 26.0
Tibble (contd)
- define a tibble row-by-row with
tribble():
tribble(
~x, ~y, ~z,
"a", 2, 3.6,
"b", 1, 8.5
)
## # A tibble: 2 x 3
## x y z
## <chr> <dbl> <dbl>
## 1 a 2.00 3.60
## 2 b 1.00 8.50
Dplyr
dplyris a grammar of data manipulation, providing a consistent set of verbs to solve the most common data manipulation challenges:mutate()adds new variables that are functions of existing variablesselect()picks variables based on their names.filter()picks cases based on their values.summarise()reduces multiple values down to a single summary.arrange()changes the ordering of the rows.
- These all combine naturally with
group_by()which allows you to perform any operation "by group"
Example
daily %>% head
## # A tibble: 6 x 15
## Type Name Department PI PIDept Status Day SerialJ Serial
## <chr> <chr> <chr> <chr> <chr> <chr> <date> <chr> <chr>
## 1 User " use… " EN/Mecha… " pi… " Mec… " Gr… 2017-10-01 " 0" " 0.0…
## 2 User " use… " Mechanic… " pi… " Mec… " Gu… 2017-10-01 " 0" " 0.0…
## 3 User " use… " EN/Mecha… " pi… " Mec… " Gr… 2017-10-01 " 0" " 0.0…
## 4 User " use… " EN/Mecha… " pi… " Mec… " Gr… 2017-10-01 " 0" " 0.0…
## 5 User " use… " EN/Mecha… " pi… " Mec… " Gr… 2017-10-01 " 0" " 0.0…
## 6 User " use… " EN/Mecha… " pi… " Mec… " Gr… 2017-10-01 " 0" " 0.0…
## # ... with 6 more variables: SingleJ <chr>, Single <chr>, MultiJ <chr>,
## # Multi <chr>, TotalJ <chr>, Total <chr>
# Number of core hours available per month for AY 2016-17
# Oct 1, 2016: Initial launch with 780 cpu
# Mar 15, 2017: Added 192 cpus
# May 1, 2017: Added 312 cpus
# Total Available at end of AY 2016-17: 1284 cpus
# Nov 15, 2017: Added 16 cpus (himem node)
# Apr 2, 2018: Added 252 cpus
# Aug 31, 2018: Added 72 cpus
# Total Available at end of AY 2017-18: 1624 cpus
# Nov 12, 2018: Added 72 cpus
# Dec 1, 2018: Added 24 cpus
# Jan 2, 2019: Added 432 cpus
# Total Available in AY 2018-19: 2152 cpus
ay1617su <- c(580320.00,561600.00,580320.00,580320.00,524160.00,580320.00,699840.00,955296.00,924480.00,955296.00,955296.00,924480.00)
ay1718su <- c(955296.00,924480.00,967200.00,967200.00,873600.00,967200.00,1117440.00, 1154688.00,1117440.00,1154688.00,1155480.00,1169280.00)
Example
monthly <- daily %>%
group_by(Month=floor_date(as.Date(Day), "month"),Name,Department,PI,PIDept,Status) %>%
summarize(Serial=sum(as.double(Serial)),
Single=sum(as.double(Single)),
Multi=sum(as.double(Multi)),
Total=sum(as.double(Total)),
SerialJ=sum(as.double(SerialJ)),
SingleJ=sum(as.double(SingleJ)),
MultiJ=sum(as.double(MultiJ)),
TotalJ=sum(as.double(TotalJ)))
monthly %>% head
## # A tibble: 6 x 14
## # Groups: Month, Name, Department, PI, PIDept [6]
## Month Name Department PI PIDept Status Serial Single Multi
## <date> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl>
## 1 2016-10-01 user0… AS/Physics… pi009 Physics Gradua… 0 18542 0
## 2 2016-10-01 user1… LTS pi016 LTS Facult… 0 817 6.38e³
## 3 2016-10-01 user1… AS/Biochem… pi024 Biolog… Gradua… 0 0 1.10e⁵
## 4 2016-10-01 user1… Biological… pi024 Biolog… Facult… 0 0 1.42e⁴
## 5 2016-10-01 user1… AS/Biochem… pi024 Biolog… Gradua… 0 14442 1.38e⁵
## 6 2016-11-01 user1… LTS pi016 LTS Facult… 0 334 3.11e⁰
## # ... with 5 more variables: Total <dbl>, SerialJ <dbl>, SingleJ <dbl>,
## # MultiJ <dbl>, TotalJ <dbl>
Sol usage per month
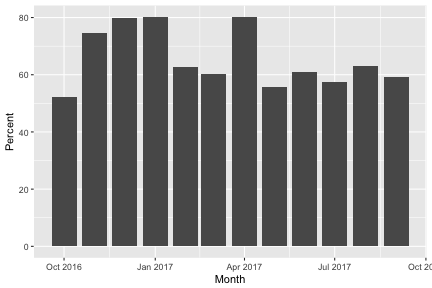
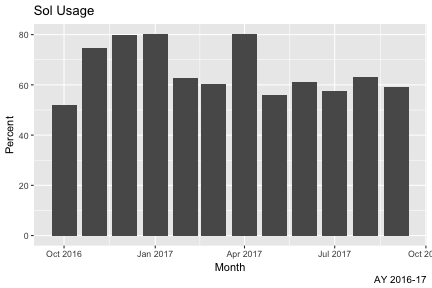
monthly %>%
group_by(Month) %>%
summarize(Total=round(sum(as.double(Total)),2),Jobs=round(sum(as.double(TotalJ)))) %>%
mutate(Available=ay1617su,Unused=Available-Total,Percent=round(Total/Available*100,2)) -> monthlyusage
monthlyusage
## # A tibble: 12 x 6
## Month Total Jobs Available Unused Percent
## <date> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2016-10-01 302612. 582. 580320. 277708. 52.2
## 2 2016-11-01 419350. 24078. 561600. 142250. 74.7
## 3 2016-12-01 463780. 21395. 580320. 116540. 79.9
## 4 2017-01-01 465491. 7004. 580320. 114829. 80.2
## 5 2017-02-01 328076. 10998. 524160. 196084. 62.6
## 6 2017-03-01 349473. 11273. 580320. 230847. 60.2
## 7 2017-04-01 560704. 6333. 699840. 139136. 80.1
## 8 2017-05-01 533431. 21804. 955296. 421865. 55.8
## 9 2017-06-01 564697. 5441. 924480. 359783. 61.1
## 10 2017-07-01 549729. 19893. 955296. 405567. 57.6
## 11 2017-08-01 602890. 45746. 955296. 352406. 63.1
## 12 2017-09-01 548241. 31076. 924480. 376239. 59.3
Sol usage per PI's Department
library(knitr)
monthly %>%
group_by(PIDept) %>%
summarize(Total=round(sum(as.double(Total)),2),Jobs=round(sum(as.double(TotalJ)))) %>%
kable
| PIDept | Total | Jobs |
|---|---|---|
| Biological Sciences | 3178662.31 | 187842 |
| Chemical and Biomolecular Engr | 1075569.60 | 4516 |
| Chemical Engineering | 44747.30 | 3257 |
| Chemistry | 24950.47 | 1185 |
| Civil and Environmental Engr | 32294.69 | 1320 |
| Computer Sci and Engineering | 21673.44 | 463 |
| LTS | 52167.27 | 2791 |
| Mathematics | 20104.04 | 32 |
| Mechanical Engr and Mechanics | 871391.19 | 3351 |
| Physics | 366912.48 | 866 |
Sol usage by user's department or major
monthly %>%
group_by(Department) %>%
summarize(Serial=round(sum(as.double(Serial))),SMP=round(sum(as.double(Single))),DMP=round(sum(as.double(Multi))),Total=round(sum(as.double(Total)),2),
Jobs=round(sum(as.double(TotalJ)))) %>% arrange(desc(Total)) %>% kable
| Department | Serial | SMP | DMP | Total | Jobs |
|---|---|---|---|---|---|
| AS/Biochemistry (CAS) | 225602 | 365083 | 2077618 | 2668302.97 | 76047 |
| EN/Chemical Engineering | 20748 | 346296 | 286186 | 653229.90 | 5173 |
| EN/Mechanical Engineering | 0 | 321597 | 300579 | 622176.07 | 2320 |
| Biological Sciences | 172290 | 73569 | 209243 | 455101.83 | 86726 |
| Chemical and Biomolecular Engr | 2663 | 272962 | 175245 | 450870.03 | 1483 |
| AS/Physics (AS) | 340 | 159440 | 210693 | 370473.39 | 987 |
| EN/ | 4285 | 41891 | 114646 | 160821.65 | 598 |
| Mechanical Engr and Mechanics | 61 | 16614 | 79776 | 96451.62 | 865 |
| LTS | 325 | 9051 | 42415 | 51790.48 | 2707 |
| Civil and Environmental Engr | 161 | 32134 | 0 | 32294.69 | 1320 |
| Chemistry | 0 | 24950 | 0 | 24950.47 | 1185 |
| Mathematics | 0 | 20104 | 0 | 20104.04 | 32 |
| IC/Computer Science & Business | 0 | 7506 | 11559 | 19065.68 | 188 |
| Computer Sci and Engineering | 10759 | 7725 | 0 | 18483.86 | 24550 |
| AS/Behavioral Neuroscience | 0 | 15132 | 2087 | 17219.65 | 418 |
| EN/Computer Science (EN) | 13 | 15921 | 0 | 15934.02 | 392 |
| EN/Bioengineering | 0 | 2032 | 6512 | 8544.27 | 25 |
| Chemical Engineering | 0 | 2170 | 0 | 2170.35 | 357 |
| AS/Chemistry (AS) | 0 | 345 | 137 | 482.21 | 239 |
| EN/Computer Engineering | 5 | 0 | 0 | 5.44 | 6 |
| EN/Structural Engineering | 0 | 0 | 0 | 0.17 | 2 |
| AS/Mathematics | 0 | 0 | 0 | 0.00 | 3 |
Need code for creating LaTeX documents
library(xtable)
monthly %>%
group_by(Department) %>%
summarize(Serial=round(sum(as.double(Serial))),SMP=round(sum(as.double(Single))),DMP=round(sum(as.double(Multi))),Total=round(sum(as.double(Total)),2),
Jobs=round(sum(as.double(TotalJ)))) %>% arrange(desc(Total)) %>% xtable
## % latex table generated in R 3.4.3 by xtable 1.8-2 package
## % Thu Feb 28 10:10:52 2019
## \begin{table}[ht]
## \centering
## \begin{tabular}{rlrrrrr}
## \hline
## & Department & Serial & SMP & DMP & Total & Jobs \\
## \hline
## 1 & AS/Biochemistry (CAS) & 225602.00 & 365083.00 & 2077618.00 & 2668302.97 & 76047.00 \\
## 2 & EN/Chemical Engineering & 20748.00 & 346296.00 & 286186.00 & 653229.90 & 5173.00 \\
## 3 & EN/Mechanical Engineering & 0.00 & 321597.00 & 300579.00 & 622176.07 & 2320.00 \\
## 4 & Biological Sciences & 172290.00 & 73569.00 & 209243.00 & 455101.83 & 86726.00 \\
## 5 & Chemical and Biomolecular Engr & 2663.00 & 272962.00 & 175245.00 & 450870.03 & 1483.00 \\
## 6 & AS/Physics (AS) & 340.00 & 159440.00 & 210693.00 & 370473.39 & 987.00 \\
## 7 & EN/ & 4285.00 & 41891.00 & 114646.00 & 160821.65 & 598.00 \\
## 8 & Mechanical Engr and Mechanics & 61.00 & 16614.00 & 79776.00 & 96451.62 & 865.00 \\
## 9 & LTS & 325.00 & 9051.00 & 42415.00 & 51790.48 & 2707.00 \\
## 10 & Civil and Environmental Engr & 161.00 & 32134.00 & 0.00 & 32294.69 & 1320.00 \\
## 11 & Chemistry & 0.00 & 24950.00 & 0.00 & 24950.47 & 1185.00 \\
## 12 & Mathematics & 0.00 & 20104.00 & 0.00 & 20104.04 & 32.00 \\
## 13 & IC/Computer Science \& Business & 0.00 & 7506.00 & 11559.00 & 19065.68 & 188.00 \\
## 14 & Computer Sci and Engineering & 10759.00 & 7725.00 & 0.00 & 18483.86 & 24550.00 \\
## 15 & AS/Behavioral Neuroscience & 0.00 & 15132.00 & 2087.00 & 17219.65 & 418.00 \\
## 16 & EN/Computer Science (EN) & 13.00 & 15921.00 & 0.00 & 15934.02 & 392.00 \\
## 17 & EN/Bioengineering & 0.00 & 2032.00 & 6512.00 & 8544.27 & 25.00 \\
## 18 & Chemical Engineering & 0.00 & 2170.00 & 0.00 & 2170.35 & 357.00 \\
## 19 & AS/Chemistry (AS) & 0.00 & 345.00 & 137.00 & 482.21 & 239.00 \\
## 20 & EN/Computer Engineering & 5.00 & 0.00 & 0.00 & 5.44 & 6.00 \\
## 21 & EN/Structural Engineering & 0.00 & 0.00 & 0.00 & 0.17 & 2.00 \\
## 22 & AS/Mathematics & 0.00 & 0.00 & 0.00 & 0.00 & 3.00 \\
## \hline
## \end{tabular}
## \end{table}
Sol usage by user affiliation
monthly %>%
group_by(Status) %>%
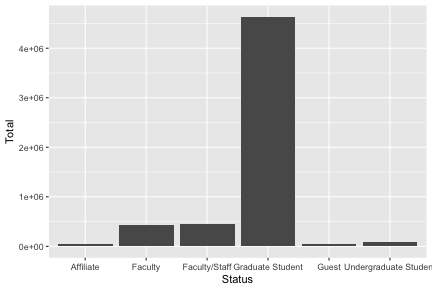
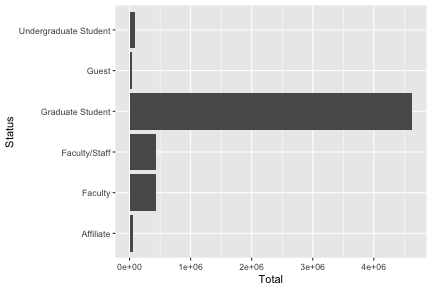
summarize(Total=round(sum(as.double(Total)),2)) -> monthlystatus
monthlystatus
## # A tibble: 6 x 2
## Status Total
## <chr> <dbl>
## 1 Affiliate 49513.
## 2 Faculty 436898.
## 3 Faculty/Staff 441351.
## 4 Graduate Student 4631676.
## 5 Guest 40979.
## 6 Undergraduate Student 88055.
Tidyr
- The goal of
tidyris to help you create tidy data. - Tidy data is data where:
- Each variable is in a column.
- Each observation is a row.
- Each value is a cell.
- Tidy data describes a standard way of storing data that is used wherever
possible throughout the
tidyverse. If you ensure that your data is tidy, you’ll spend less timing fighting with the tools and more time working on your analysis.
two fundamental verbs of data tidying:
gather()takes multiple columns, and gathers them into key-value pairsspread(). takes two columns (key & value) and spreads in to multiple columns
Example
daily %>%
filter(as.Date(Day) >= "2017-02-01" & as.Date(Day) <= "2017-03-01") %>%
select(Day,Name,Department,PI,PIDept,Serial,Single,Multi) %>%
gather(JobType,Usage,Serial:Multi) %>%
filter(as.double(Usage) > 100 ) -> tmp
tmp %>% arrange(Usage) %>%
kable
| Day | Name | Department | PI | PIDept | JobType | Usage |
|---|---|---|---|---|---|---|
| 2017-02-20 | user140 | Civil and Environmental Engr | pi020 | Civil and Environmental Engr | Single | 104.1611 |
| 2017-02-07 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 104.7500 |
| 2017-02-27 | user163 | Biological Sciences | pi024 | Biological Sciences | Serial | 105.4464 |
| 2017-02-15 | user023 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 107.6944 |
| 2017-02-11 | user027 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 108.8500 |
| 2017-02-15 | user035 | AS/Behavioral Neuroscience | pi005 | Chemical Engineering | Single | 114.1444 |
| 2017-02-04 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 114.2278 |
| 2017-02-20 | user021 | AS/Physics (AS) | pi005 | Chemical Engineering | Single | 115.1944 |
| 2017-02-15 | user128 | LTS | pi016 | LTS | Single | 115.4333 |
| 2017-02-16 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Single | 118.3239 |
| 2017-02-28 | user021 | AS/Physics (AS) | pi005 | Chemical Engineering | Single | 120.1389 |
| 2017-02-17 | user036 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 121.1556 |
| 2017-02-16 | user036 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 122.4833 |
| 2017-02-17 | user022 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 126.5444 |
| 2017-02-02 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 129.8619 |
| 2017-02-15 | user036 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 130.0778 |
| 2017-02-21 | user021 | AS/Physics (AS) | pi005 | Chemical Engineering | Single | 130.3667 |
| 2017-02-18 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 133.4642 |
| 2017-02-16 | user038 | AS/Chemistry (AS) | pi005 | Chemical Engineering | Multi | 133.8667 |
| 2017-02-22 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 134.4111 |
| 2017-02-16 | user038 | AS/Chemistry (AS) | pi005 | Chemical Engineering | Single | 136.4111 |
| 2017-02-02 | user138 | Chemistry | pi018 | Chemistry | Single | 138.5944 |
| 2017-02-15 | user032 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 139.7722 |
| 2017-02-16 | user006 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 140.1167 |
| 2017-02-19 | user021 | AS/Physics (AS) | pi005 | Chemical Engineering | Single | 140.7222 |
| 2017-02-19 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 141.3272 |
| 2017-02-15 | user024 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 142.1389 |
| 2017-02-23 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Multi | 142.6778 |
| 2017-03-01 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 145.2944 |
| 2017-02-01 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 145.3694 |
| 2017-02-14 | user027 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 146.3333 |
| 2017-02-24 | user023 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 146.4556 |
| 2017-02-22 | user021 | AS/Physics (AS) | pi005 | Chemical Engineering | Single | 148.0778 |
| 2017-02-08 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 150.2964 |
| 2017-02-22 | user023 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 156.0111 |
| 2017-02-14 | user023 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 158.6667 |
| 2017-02-15 | user027 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 159.6167 |
| 2017-02-04 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 164.4814 |
| 2017-02-11 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 166.4300 |
| 2017-02-06 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 170.2172 |
| 2017-03-01 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 172.6736 |
| 2017-02-19 | user159 | Biological Sciences | pi024 | Biological Sciences | Serial | 175.5294 |
| 2017-02-11 | user138 | Chemistry | pi018 | Chemistry | Single | 178.8278 |
| 2017-02-10 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 183.3172 |
| 2017-02-13 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 188.1469 |
| 2017-02-12 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 197.7642 |
| 2017-02-28 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 199.3139 |
| 2017-02-24 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Single | 206.0994 |
| 2017-02-24 | user030 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 206.3444 |
| 2017-02-20 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 206.3906 |
| 2017-02-21 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Single | 207.0111 |
| 2017-02-03 | user138 | Chemistry | pi018 | Chemistry | Single | 212.6167 |
| 2017-02-21 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 216.3067 |
| 2017-02-03 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 217.5011 |
| 2017-02-27 | user159 | Biological Sciences | pi024 | Biological Sciences | Serial | 221.7922 |
| 2017-02-17 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 222.0128 |
| 2017-02-14 | user024 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 222.6111 |
| 2017-02-27 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 224.1025 |
| 2017-02-18 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Single | 224.1867 |
| 2017-02-04 | user138 | Chemistry | pi018 | Chemistry | Single | 228.2333 |
| 2017-02-22 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 231.3806 |
| 2017-02-24 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 232.4611 |
| 2017-02-15 | user022 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 233.7667 |
| 2017-02-14 | user006 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 235.3556 |
| 2017-02-14 | user022 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 237.0889 |
| 2017-02-17 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 240.7922 |
| 2017-02-23 | user177 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 243.8686 |
| 2017-02-25 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Multi | 254.4778 |
| 2017-02-09 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 255.9514 |
| 2017-02-18 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 265.7314 |
| 2017-02-26 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 266.3714 |
| 2017-02-16 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 277.4244 |
| 2017-02-15 | user021 | AS/Physics (AS) | pi005 | Chemical Engineering | Single | 280.4333 |
| 2017-02-14 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 283.6800 |
| 2017-02-07 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 288.1375 |
| 2017-02-24 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 290.8047 |
| 2017-02-05 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 294.1478 |
| 2017-02-06 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 297.9114 |
| 2017-02-04 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Multi | 300.0778 |
| 2017-02-23 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 307.9611 |
| 2017-02-16 | user022 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 326.0611 |
| 2017-02-12 | user138 | Chemistry | pi018 | Chemistry | Single | 326.1778 |
| 2017-02-25 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 366.0944 |
| 2017-02-01 | user138 | Chemistry | pi018 | Chemistry | Single | 372.3222 |
| 2017-02-25 | user159 | Biological Sciences | pi024 | Biological Sciences | Serial | 408.5003 |
| 2017-02-17 | user022 | EN/Chemical Engineering | pi005 | Chemical Engineering | Multi | 410.5556 |
| 2017-02-22 | user163 | Biological Sciences | pi024 | Biological Sciences | Serial | 414.5008 |
| 2017-02-23 | user023 | EN/Chemical Engineering | pi005 | Chemical Engineering | Single | 425.0167 |
| 2017-02-14 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Single | 425.1778 |
| 2017-02-16 | user031 | EN/ | pi005 | Chemical Engineering | Single | 430.6056 |
| 2017-02-15 | user031 | EN/ | pi005 | Chemical Engineering | Single | 453.6500 |
| 2017-02-28 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 460.2444 |
| 2017-02-20 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Single | 465.7378 |
| 2017-02-22 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Single | 503.2222 |
| 2017-02-23 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Single | 521.5822 |
| 2017-02-12 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Single | 541.8711 |
| 2017-02-21 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Single | 553.0756 |
| 2017-02-11 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Single | 618.3822 |
| 2017-02-13 | user138 | Chemistry | pi018 | Chemistry | Single | 640.1889 |
| 2017-02-22 | user159 | Biological Sciences | pi024 | Biological Sciences | Serial | 661.2825 |
| 2017-02-23 | user009 | Chemical and Biomolecular Engr | pi002 | Chemical and Biomolecular Engr | Single | 692.3056 |
| 2017-02-13 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Single | 711.0267 |
| 2017-02-01 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 720.4808 |
| 2017-02-10 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Single | 735.6622 |
| 2017-03-01 | user031 | EN/ | pi005 | Chemical Engineering | Single | 741.6833 |
| 2017-02-17 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Single | 747.2578 |
| 2017-02-14 | user128 | LTS | pi016 | LTS | Single | 750.6067 |
| 2017-02-09 | user122 | AS/Physics (AS) | pi013 | Physics | Multi | 752.5500 |
| 2017-02-21 | user006 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 778.3556 |
| 2017-02-05 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 780.2081 |
| 2017-02-14 | user138 | Chemistry | pi018 | Chemistry | Single | 802.0389 |
| 2017-02-02 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 819.1667 |
| 2017-02-03 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | Single | 854.3111 |
| 2017-02-22 | user006 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 887.0833 |
| 2017-02-08 | user163 | Biological Sciences | pi024 | Biological Sciences | Serial | 895.3944 |
| 2017-03-01 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | Single | 928.9500 |
| 2017-02-16 | user021 | AS/Physics (AS) | pi005 | Chemical Engineering | Single | 931.9722 |
| 2017-02-25 | user031 | EN/ | pi005 | Chemical Engineering | Single | 960.0222 |
| 2017-02-09 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 1021.2722 |
| 2017-02-10 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Multi | 1051.5222 |
| 2017-02-19 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 1110.0111 |
| 2017-02-25 | user009 | Chemical and Biomolecular Engr | pi002 | Chemical and Biomolecular Engr | Single | 1130.2333 |
| 2017-02-22 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 1227.3067 |
| 2017-02-07 | user122 | AS/Physics (AS) | pi013 | Physics | Multi | 1246.4833 |
| 2017-02-04 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 1357.3228 |
| 2017-02-26 | user009 | Chemical and Biomolecular Engr | pi002 | Chemical and Biomolecular Engr | Single | 1362.2833 |
| 2017-02-16 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 1396.8778 |
| 2017-02-24 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | Single | 1432.2000 |
| 2017-02-01 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | Single | 1440.0500 |
| 2017-02-02 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | Single | 1440.0500 |
| 2017-02-06 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | Single | 1440.1778 |
| 2017-02-10 | user122 | AS/Physics (AS) | pi013 | Physics | Multi | 1440.3667 |
| 2017-02-10 | user163 | Biological Sciences | pi024 | Biological Sciences | Serial | 1467.6819 |
| 2017-02-24 | user031 | EN/ | pi005 | Chemical Engineering | Single | 1478.1056 |
| 2017-03-01 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Multi | 1573.7667 |
| 2017-02-26 | user159 | Biological Sciences | pi024 | Biological Sciences | Serial | 1633.3575 |
| 2017-03-01 | user009 | Chemical and Biomolecular Engr | pi002 | Chemical and Biomolecular Engr | Single | 1643.1611 |
| 2017-02-14 | user122 | AS/Physics (AS) | pi013 | Physics | Multi | 1832.3800 |
| 2017-02-02 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 2142.4653 |
| 2017-02-09 | user163 | Biological Sciences | pi024 | Biological Sciences | Serial | 2379.7086 |
| 2017-02-09 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 2457.0700 |
| 2017-02-06 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Multi | 2566.5444 |
| 2017-02-03 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Serial | 2616.1383 |
| 2017-02-22 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Multi | 2721.7333 |
| 2017-02-27 | user006 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 2829.9611 |
| 2017-02-09 | user070 | AS/Physics (AS) | pi009 | Physics | Single | 2840.2722 |
| 2017-02-24 | user006 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 2863.0389 |
| 2017-02-16 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | Multi | 2866.4556 |
| 2017-02-09 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | Multi | 2880.2111 |
| 2017-02-04 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | Multi | 2880.2556 |
| 2017-02-08 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | Multi | 2880.3222 |
| 2017-02-06 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | Multi | 2880.3667 |
| 2017-02-20 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | Multi | 2880.6889 |
| 2017-03-01 | user159 | Biological Sciences | pi024 | Biological Sciences | Serial | 3317.4100 |
| 2017-02-28 | user009 | Chemical and Biomolecular Engr | pi002 | Chemical and Biomolecular Engr | Single | 3465.9111 |
| 2017-02-07 | user127 | AS/Behavioral Neuroscience | pi015 | Biological Sciences | Single | 3725.7061 |
| 2017-02-23 | user163 | Biological Sciences | pi024 | Biological Sciences | Serial | 3773.1967 |
| 2017-02-28 | user159 | Biological Sciences | pi024 | Biological Sciences | Serial | 3888.3147 |
| 2017-02-18 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 4015.0000 |
| 2017-02-20 | user006 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 4075.2889 |
| 2017-02-17 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 4101.8000 |
| 2017-02-13 | user006 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Single | 4290.1278 |
| 2017-02-03 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Multi | 4899.0222 |
| 2017-02-27 | user009 | Chemical and Biomolecular Engr | pi002 | Chemical and Biomolecular Engr | Single | 4922.7056 |
| 2017-02-01 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 5264.1233 |
| 2017-02-03 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 5610.3300 |
| 2017-02-01 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | Multi | 5715.8889 |
| 2017-02-12 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 6794.1900 |
| 2017-02-10 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 6893.1000 |
| 2017-02-23 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 6898.0633 |
| 2017-02-05 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 6959.4800 |
| 2017-02-13 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 7023.4200 |
| 2017-02-11 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 7177.8600 |
| 2017-02-04 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 7547.7600 |
| 2017-02-24 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 7622.2667 |
| 2017-02-06 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 8071.8733 |
| 2017-03-01 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 8678.0833 |
| 2017-02-08 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 8935.5200 |
| 2017-02-27 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 9322.2800 |
| 2017-02-28 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 9503.2300 |
| 2017-02-25 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 9641.4633 |
| 2017-02-26 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 9705.6667 |
| 2017-02-14 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 10380.3300 |
| 2017-02-02 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 12405.5800 |
| 2017-02-07 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 12804.9900 |
| 2017-02-20 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | Multi | 17280.4000 |
Example
tmp %>% arrange(Usage) %>%
spread(JobType,Usage,fill = 0.0) %>%
kable
| Day | Name | Department | PI | PIDept | Multi | Serial | Single |
|---|---|---|---|---|---|---|---|
| 2017-02-01 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 5715.8889 | 0.0000 | 0.0000 |
| 2017-02-01 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 1440.0500 |
| 2017-02-01 | user138 | Chemistry | pi018 | Chemistry | 0.0000 | 0.0000 | 372.3222 |
| 2017-02-01 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 5264.1233 | 0.0000 | 0.0000 |
| 2017-02-01 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 720.4808 | 0.0000 |
| 2017-02-01 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 145.3694 | 0.0000 |
| 2017-02-02 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 1440.0500 |
| 2017-02-02 | user138 | Chemistry | pi018 | Chemistry | 0.0000 | 0.0000 | 138.5944 |
| 2017-02-02 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 12405.5800 | 819.1667 | 0.0000 |
| 2017-02-02 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 2142.4653 | 0.0000 |
| 2017-02-02 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 129.8619 | 0.0000 |
| 2017-02-03 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 4899.0222 | 0.0000 | 0.0000 |
| 2017-02-03 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 854.3111 |
| 2017-02-03 | user138 | Chemistry | pi018 | Chemistry | 0.0000 | 0.0000 | 212.6167 |
| 2017-02-03 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 5610.3300 | 0.0000 | 0.0000 |
| 2017-02-03 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 2616.1383 | 0.0000 |
| 2017-02-03 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 217.5011 | 0.0000 |
| 2017-02-04 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 300.0778 | 0.0000 | 114.2278 |
| 2017-02-04 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | 2880.2556 | 0.0000 | 0.0000 |
| 2017-02-04 | user138 | Chemistry | pi018 | Chemistry | 0.0000 | 0.0000 | 228.2333 |
| 2017-02-04 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 7547.7600 | 0.0000 | 0.0000 |
| 2017-02-04 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 1357.3228 | 0.0000 |
| 2017-02-04 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 164.4814 | 0.0000 |
| 2017-02-05 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 6959.4800 | 0.0000 | 0.0000 |
| 2017-02-05 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 780.2081 | 0.0000 |
| 2017-02-05 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 294.1478 | 0.0000 |
| 2017-02-06 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 2566.5444 | 0.0000 | 0.0000 |
| 2017-02-06 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | 2880.3667 | 0.0000 | 1440.1778 |
| 2017-02-06 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 8071.8733 | 0.0000 | 0.0000 |
| 2017-02-06 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 297.9114 | 0.0000 |
| 2017-02-06 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 170.2172 | 0.0000 |
| 2017-02-07 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 104.7500 |
| 2017-02-07 | user122 | AS/Physics (AS) | pi013 | Physics | 1246.4833 | 0.0000 | 0.0000 |
| 2017-02-07 | user127 | AS/Behavioral Neuroscience | pi015 | Biological Sciences | 0.0000 | 0.0000 | 3725.7061 |
| 2017-02-07 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 12804.9900 | 0.0000 | 0.0000 |
| 2017-02-07 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 288.1375 | 0.0000 |
| 2017-02-08 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | 2880.3222 | 0.0000 | 0.0000 |
| 2017-02-08 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 8935.5200 | 0.0000 | 0.0000 |
| 2017-02-08 | user163 | Biological Sciences | pi024 | Biological Sciences | 0.0000 | 895.3944 | 0.0000 |
| 2017-02-08 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 150.2964 | 0.0000 |
| 2017-02-09 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 1021.2722 |
| 2017-02-09 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | 2880.2111 | 0.0000 | 0.0000 |
| 2017-02-09 | user070 | AS/Physics (AS) | pi009 | Physics | 0.0000 | 0.0000 | 2840.2722 |
| 2017-02-09 | user122 | AS/Physics (AS) | pi013 | Physics | 752.5500 | 0.0000 | 0.0000 |
| 2017-02-09 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 2457.0700 | 0.0000 | 0.0000 |
| 2017-02-09 | user163 | Biological Sciences | pi024 | Biological Sciences | 0.0000 | 2379.7086 | 0.0000 |
| 2017-02-09 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 255.9514 | 0.0000 |
| 2017-02-10 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 1051.5222 | 0.0000 | 0.0000 |
| 2017-02-10 | user122 | AS/Physics (AS) | pi013 | Physics | 1440.3667 | 0.0000 | 0.0000 |
| 2017-02-10 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 6893.1000 | 0.0000 | 0.0000 |
| 2017-02-10 | user163 | Biological Sciences | pi024 | Biological Sciences | 0.0000 | 1467.6819 | 0.0000 |
| 2017-02-10 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 183.3172 | 0.0000 |
| 2017-02-10 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 0.0000 | 735.6622 |
| 2017-02-11 | user027 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 108.8500 |
| 2017-02-11 | user138 | Chemistry | pi018 | Chemistry | 0.0000 | 0.0000 | 178.8278 |
| 2017-02-11 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 7177.8600 | 0.0000 | 0.0000 |
| 2017-02-11 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 166.4300 | 0.0000 |
| 2017-02-11 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 0.0000 | 618.3822 |
| 2017-02-12 | user138 | Chemistry | pi018 | Chemistry | 0.0000 | 0.0000 | 326.1778 |
| 2017-02-12 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 6794.1900 | 0.0000 | 0.0000 |
| 2017-02-12 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 197.7642 | 0.0000 |
| 2017-02-12 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 0.0000 | 541.8711 |
| 2017-02-13 | user006 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 4290.1278 |
| 2017-02-13 | user138 | Chemistry | pi018 | Chemistry | 0.0000 | 0.0000 | 640.1889 |
| 2017-02-13 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 7023.4200 | 0.0000 | 0.0000 |
| 2017-02-13 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 188.1469 | 0.0000 |
| 2017-02-13 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 0.0000 | 711.0267 |
| 2017-02-14 | user006 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 235.3556 |
| 2017-02-14 | user022 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 237.0889 |
| 2017-02-14 | user023 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 158.6667 |
| 2017-02-14 | user024 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 222.6111 |
| 2017-02-14 | user027 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 146.3333 |
| 2017-02-14 | user122 | AS/Physics (AS) | pi013 | Physics | 1832.3800 | 0.0000 | 0.0000 |
| 2017-02-14 | user128 | LTS | pi016 | LTS | 0.0000 | 0.0000 | 750.6067 |
| 2017-02-14 | user138 | Chemistry | pi018 | Chemistry | 0.0000 | 0.0000 | 802.0389 |
| 2017-02-14 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 10380.3300 | 0.0000 | 0.0000 |
| 2017-02-14 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 283.6800 | 0.0000 |
| 2017-02-14 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 0.0000 | 425.1778 |
| 2017-02-15 | user021 | AS/Physics (AS) | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 280.4333 |
| 2017-02-15 | user022 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 233.7667 |
| 2017-02-15 | user023 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 107.6944 |
| 2017-02-15 | user024 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 142.1389 |
| 2017-02-15 | user027 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 159.6167 |
| 2017-02-15 | user031 | EN/ | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 453.6500 |
| 2017-02-15 | user032 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 139.7722 |
| 2017-02-15 | user035 | AS/Behavioral Neuroscience | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 114.1444 |
| 2017-02-15 | user036 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 130.0778 |
| 2017-02-15 | user128 | LTS | pi016 | LTS | 0.0000 | 0.0000 | 115.4333 |
| 2017-02-16 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 1396.8778 |
| 2017-02-16 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | 2866.4556 | 0.0000 | 0.0000 |
| 2017-02-16 | user006 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 140.1167 |
| 2017-02-16 | user021 | AS/Physics (AS) | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 931.9722 |
| 2017-02-16 | user022 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 326.0611 |
| 2017-02-16 | user031 | EN/ | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 430.6056 |
| 2017-02-16 | user036 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 122.4833 |
| 2017-02-16 | user038 | AS/Chemistry (AS) | pi005 | Chemical Engineering | 133.8667 | 0.0000 | 136.4111 |
| 2017-02-16 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 277.4244 | 118.3239 |
| 2017-02-17 | user022 | EN/Chemical Engineering | pi005 | Chemical Engineering | 410.5556 | 0.0000 | 126.5444 |
| 2017-02-17 | user036 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 121.1556 |
| 2017-02-17 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 4101.8000 | 0.0000 | 0.0000 |
| 2017-02-17 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 240.7922 | 0.0000 |
| 2017-02-17 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 222.0128 | 0.0000 |
| 2017-02-17 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 0.0000 | 747.2578 |
| 2017-02-18 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 4015.0000 | 0.0000 | 0.0000 |
| 2017-02-18 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 265.7314 | 0.0000 |
| 2017-02-18 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 133.4642 | 0.0000 |
| 2017-02-18 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 0.0000 | 224.1867 |
| 2017-02-19 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 1110.0111 |
| 2017-02-19 | user021 | AS/Physics (AS) | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 140.7222 |
| 2017-02-19 | user159 | Biological Sciences | pi024 | Biological Sciences | 0.0000 | 175.5294 | 0.0000 |
| 2017-02-19 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 141.3272 | 0.0000 |
| 2017-02-20 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | 2880.6889 | 0.0000 | 0.0000 |
| 2017-02-20 | user006 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 4075.2889 |
| 2017-02-20 | user021 | AS/Physics (AS) | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 115.1944 |
| 2017-02-20 | user140 | Civil and Environmental Engr | pi020 | Civil and Environmental Engr | 0.0000 | 0.0000 | 104.1611 |
| 2017-02-20 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 17280.4000 | 0.0000 | 0.0000 |
| 2017-02-20 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 206.3906 | 0.0000 |
| 2017-02-20 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 0.0000 | 465.7378 |
| 2017-02-21 | user006 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 778.3556 |
| 2017-02-21 | user021 | AS/Physics (AS) | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 130.3667 |
| 2017-02-21 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 0.0000 | 207.0111 |
| 2017-02-21 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 216.3067 | 0.0000 |
| 2017-02-21 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 0.0000 | 553.0756 |
| 2017-02-22 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 2721.7333 | 0.0000 | 134.4111 |
| 2017-02-22 | user006 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 887.0833 |
| 2017-02-22 | user021 | AS/Physics (AS) | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 148.0778 |
| 2017-02-22 | user023 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 156.0111 |
| 2017-02-22 | user159 | Biological Sciences | pi024 | Biological Sciences | 0.0000 | 661.2825 | 0.0000 |
| 2017-02-22 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 1227.3067 | 0.0000 | 0.0000 |
| 2017-02-22 | user163 | Biological Sciences | pi024 | Biological Sciences | 0.0000 | 414.5008 | 0.0000 |
| 2017-02-22 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 231.3806 | 0.0000 |
| 2017-02-22 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 0.0000 | 503.2222 |
| 2017-02-23 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 142.6778 | 0.0000 | 307.9611 |
| 2017-02-23 | user009 | Chemical and Biomolecular Engr | pi002 | Chemical and Biomolecular Engr | 0.0000 | 0.0000 | 692.3056 |
| 2017-02-23 | user023 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 425.0167 |
| 2017-02-23 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 6898.0633 | 0.0000 | 0.0000 |
| 2017-02-23 | user163 | Biological Sciences | pi024 | Biological Sciences | 0.0000 | 3773.1967 | 0.0000 |
| 2017-02-23 | user177 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 243.8686 | 0.0000 |
| 2017-02-23 | user178 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 0.0000 | 521.5822 |
| 2017-02-24 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 232.4611 |
| 2017-02-24 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 1432.2000 |
| 2017-02-24 | user006 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 2863.0389 |
| 2017-02-24 | user023 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 146.4556 |
| 2017-02-24 | user030 | EN/Chemical Engineering | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 206.3444 |
| 2017-02-24 | user031 | EN/ | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 1478.1056 |
| 2017-02-24 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 7622.2667 | 0.0000 | 0.0000 |
| 2017-02-24 | user166 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 0.0000 | 206.0994 |
| 2017-02-24 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 290.8047 | 0.0000 |
| 2017-02-25 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 254.4778 | 0.0000 | 366.0944 |
| 2017-02-25 | user009 | Chemical and Biomolecular Engr | pi002 | Chemical and Biomolecular Engr | 0.0000 | 0.0000 | 1130.2333 |
| 2017-02-25 | user031 | EN/ | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 960.0222 |
| 2017-02-25 | user159 | Biological Sciences | pi024 | Biological Sciences | 0.0000 | 408.5003 | 0.0000 |
| 2017-02-25 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 9641.4633 | 0.0000 | 0.0000 |
| 2017-02-26 | user009 | Chemical and Biomolecular Engr | pi002 | Chemical and Biomolecular Engr | 0.0000 | 0.0000 | 1362.2833 |
| 2017-02-26 | user159 | Biological Sciences | pi024 | Biological Sciences | 0.0000 | 1633.3575 | 0.0000 |
| 2017-02-26 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 9705.6667 | 0.0000 | 0.0000 |
| 2017-02-26 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 266.3714 | 0.0000 |
| 2017-02-27 | user006 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 2829.9611 |
| 2017-02-27 | user009 | Chemical and Biomolecular Engr | pi002 | Chemical and Biomolecular Engr | 0.0000 | 0.0000 | 4922.7056 |
| 2017-02-27 | user159 | Biological Sciences | pi024 | Biological Sciences | 0.0000 | 221.7922 | 0.0000 |
| 2017-02-27 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 9322.2800 | 0.0000 | 0.0000 |
| 2017-02-27 | user163 | Biological Sciences | pi024 | Biological Sciences | 0.0000 | 105.4464 | 0.0000 |
| 2017-02-27 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 224.1025 | 0.0000 |
| 2017-02-28 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 460.2444 |
| 2017-02-28 | user009 | Chemical and Biomolecular Engr | pi002 | Chemical and Biomolecular Engr | 0.0000 | 0.0000 | 3465.9111 |
| 2017-02-28 | user021 | AS/Physics (AS) | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 120.1389 |
| 2017-02-28 | user159 | Biological Sciences | pi024 | Biological Sciences | 0.0000 | 3888.3147 | 0.0000 |
| 2017-02-28 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 9503.2300 | 0.0000 | 0.0000 |
| 2017-02-28 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 199.3139 | 0.0000 |
| 2017-03-01 | user003 | EN/Mechanical Engineering | pi001 | Mechanical Engr and Mechanics | 1573.7667 | 0.0000 | 145.2944 |
| 2017-03-01 | user005 | EN/ | pi001 | Mechanical Engr and Mechanics | 0.0000 | 0.0000 | 928.9500 |
| 2017-03-01 | user009 | Chemical and Biomolecular Engr | pi002 | Chemical and Biomolecular Engr | 0.0000 | 0.0000 | 1643.1611 |
| 2017-03-01 | user031 | EN/ | pi005 | Chemical Engineering | 0.0000 | 0.0000 | 741.6833 |
| 2017-03-01 | user159 | Biological Sciences | pi024 | Biological Sciences | 0.0000 | 3317.4100 | 0.0000 |
| 2017-03-01 | user162 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 8678.0833 | 0.0000 | 0.0000 |
| 2017-03-01 | user167 | AS/Biochemistry (CAS) | pi024 | Biological Sciences | 0.0000 | 172.6736 | 0.0000 |
Other Tidyr functions
separate(): Splitting a single variable into two
daily %>%
select(c(Department,Day,Total)) %>%
separate(Day,c("Year","Month","Day"),sep="-") -> tmp
head(tmp)
## # A tibble: 6 x 5
## Department Year Month Day Total
## <chr> <chr> <chr> <chr> <dbl>
## 1 AS/Physics (AS) 2016 10 01 2881.
## 2 AS/Biochemistry (CAS) 2016 10 01 13826.
## 3 AS/Physics (AS) 2016 10 03 2881.
## 4 AS/Biochemistry (CAS) 2016 10 03 1452.
## 5 AS/Physics (AS) 2016 10 04 0.200
## 6 LTS 2016 10 04 652.
unite(): Merging two variables into one
tmp %>%
unite(Day,c("Year","Month","Day"),sep="/") %>%
tail
## # A tibble: 6 x 3
## Department Day Total
## <chr> <chr> <dbl>
## 1 Biological Sciences 2017/09/30 0.338
## 2 AS/Biochemistry (CAS) 2017/09/30 559.
## 3 AS/Biochemistry (CAS) 2017/09/30 119.
## 4 IC/Computer Science & Business 2017/09/30 43.5
## 5 AS/Biochemistry (CAS) 2017/09/30 250.
## 6 Biological Sciences 2017/09/30 7755.
What more can be done with R?
- Data Visualization
- Data cleaning/preprocessing
- Profiling and debugging
- Regression Models
- Machine learning/Data Mining
- ···
Data Visualization
- Data visualization or data visualisation is viewed by many disciplines as a modern equivalent of visual communication.
- It involves the creation and study of the visual representation of data.
- A primary goal of data visualization is to communicate information clearly and efficiently via statistical graphics, plots and information graphics.
- Data visualization is both an art and a science.
- More details in next weeks seminar
Bar Charts
p <- monthlystatus %>%
ggplot(aes(x=Status,y=Total)) + geom_col()
p
p + coord_flip()
Bar Charts Monthly Usage
p <- monthlyusage %>%
ggplot(aes(Month,Percent)) + geom_col()
p
- Add Plot Title and Caption, and x and y labels
p + labs(title="Sol Usage", y="Percent", x="Month", caption="AY 2016-17")
Line Charts
p <- daily %>%
group_by(Day, Status=trimws(Status)) %>%
summarize(Total=round(sum(as.double(Total)),2),Jobs=round(sum(as.double(TotalJ)))) %>%
ggplot(aes(Day,Total)) + geom_line(aes(col = Status))
p
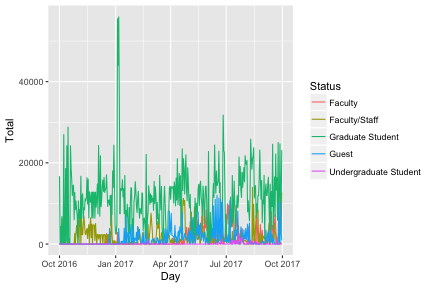
Line Charts (contd)
Plot is very busy. There are several options to clean this up.
- summarize by week or month
- take a running average or add a smoothing function
- create separate plots for each Department using facet_wrap
p + facet_wrap( ~trimws(Status), scales = "free", ncol = 2) + theme(legend.position='none')
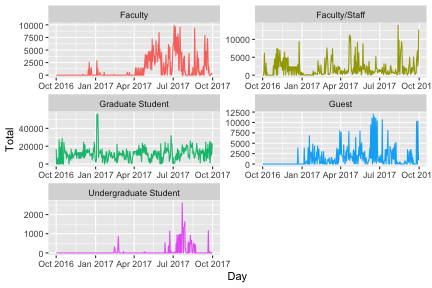
Animations
If a Picture is Worth a Thousand Words, Then Is a Video Worth a Million?
- What is an animation really?
- Just a collection of images that appear at a high frequency or frame rate.
- If you have a collection of pictures, you can convert them to gif, mpeg, or any other video format using tools like ImageMagick or ffmpeg.
- R provides tools that will convert a collection of images from plots to video provided you have one of these conversion tools.
if(!require('animation')){
install.packages('animation')
}
if(!require('gganimate')){
install.packages('animation')
}
Animations (contd)
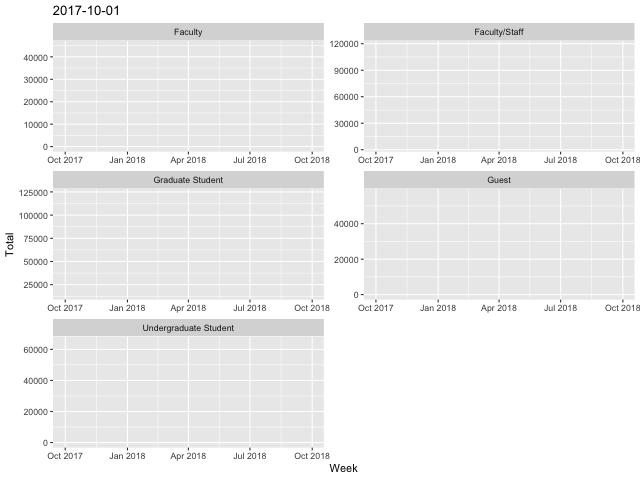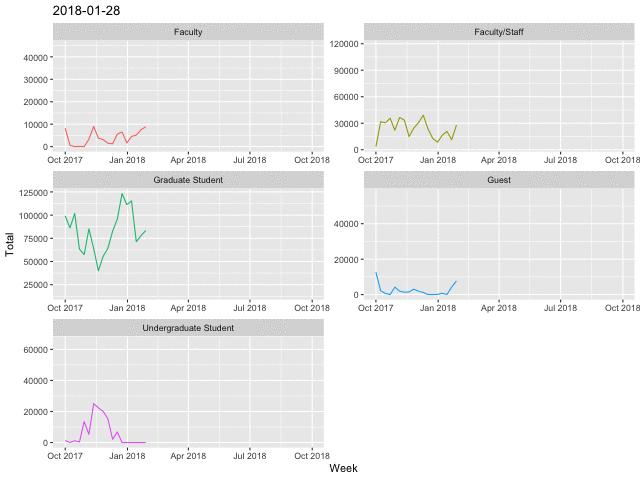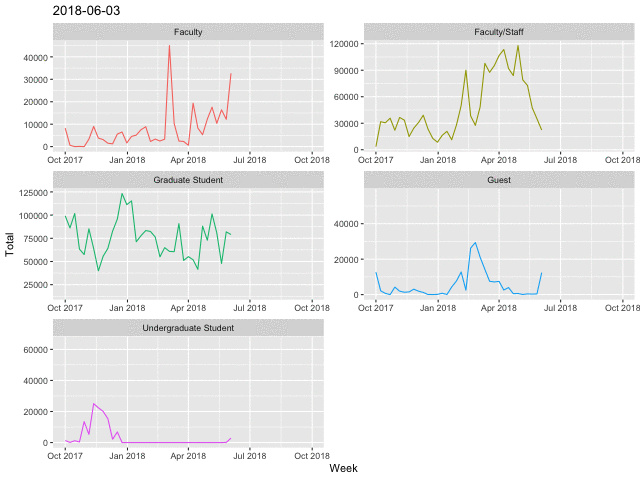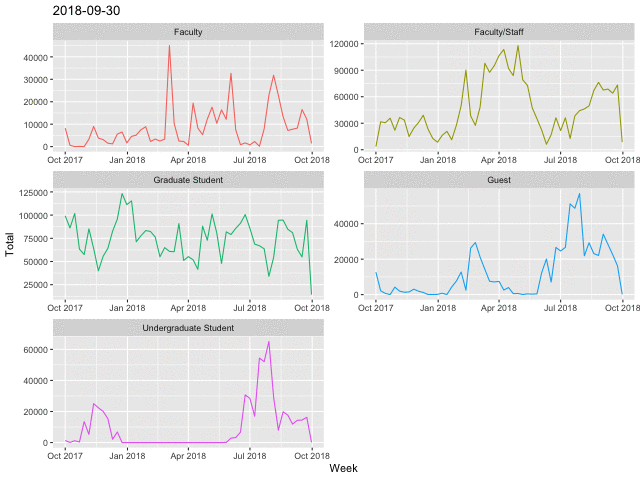
Learning R
- User documentation on CRAN
- An Introduction on R: http://cran.r-project.org/doc/manuals/r-release/R-intro.html
- Online tutorials
- Educational R packages
- Swirl: Learn R in R
- Online courses (e.g. Coursera)
Data Analysis with Reporting
- Typical data analysis workflow involves
- Obtaining the data
- Cleaning and preprocessing the data
- Analyzing the data
- Generating a report
knitris a R package that allows one to generate dynamic report by weaving R code and human readable texts together- It uses the markdown syntax
- The output can be HTML, PDF or (even) Word
slidifyis a R package that allows one to create a HTML presentation- You are now at the end of a
slidifypresentation
- You are now at the end of a
Creating presentations using Slidify
- Install the
devtoolspackage and load it
install.packages('devtools')
library(devtools)
- Install the
slidifyandslidifyLibrariespackage from github
install_git('git://github.com/ramnathv/slidify')
install_git('git://github.com/ramnathv/slidifyLibraries')
- Load the slidify library
library(slidify)
- Create a Slide desk
author("myslides")
Creating presentations using Slidify (contd)
- This will create a folder called
myslideswith files and subdirectories to create your presentationassets/css/custom.css: Create your own custom cssassets/layouts/: Don't like the default layouts, create your own in this directorylibraries: files that slidify create. Do not edit the files, copy the file to theassetsdirectory and modify it.
- To edit your presentation, edit the
index.Rmdfile using R Markdown - To create slides, in the R console run the command
slidify('index.Rmd')``
# View the presentation in a web browser
browserURL('index.html')
- Do a Google search for
slidifyto learn more and/or see example slides.
Creating presentations using Slidify (contd)
- To use my template
git clone https://gogs.cc.lehigh.edu/alp514/slidify
- Edit the index.Rmd or create a new .Rmd file
- OR just overwrite the assets folder in your
myslidesfolder with the one from the git repository you just cloned - The git repository contains a script
compile.Rthat will compile the R markdown file (for e.g. index.Rmd) into a html file (for e.g. index.html)
chmod +x compile.R
./compile.R index